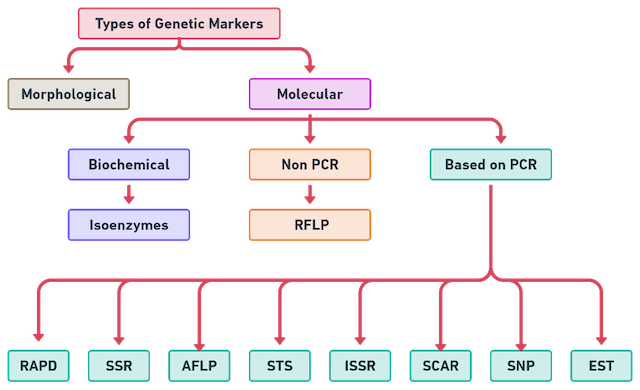
🚩 Genetics Markers
Types, Selection, Uses
- A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species.
- It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed.
- Generally, they do not represent the target genes themselves but act as ‘signs’ or ‘flags’.
- Genetic markers that are located in close proximity to genes (i.e. tightly linked) may be referred to as gene ‘tags’.
- Such markers themselves do not affect the phenotype of the trait of interest because they are located only near or ’linked’ to genes controlling the trait.
- All genetic markers occupy specific genomic positions within chromosomes (like genes) called ‘loci’ (singular ’locus’).
Types of Genetic markers
Molecular Marker
- Molecular markers are specific fragments of DNA that can be identified within the whole genome.
- Molecular markers are found at specific locations of the genome.
- They are used to ‘flag’ the position of a particular gene or the inheritance of a particular character.
- Molecular markers are
phenotypically neutral.
Important Molecular Markers
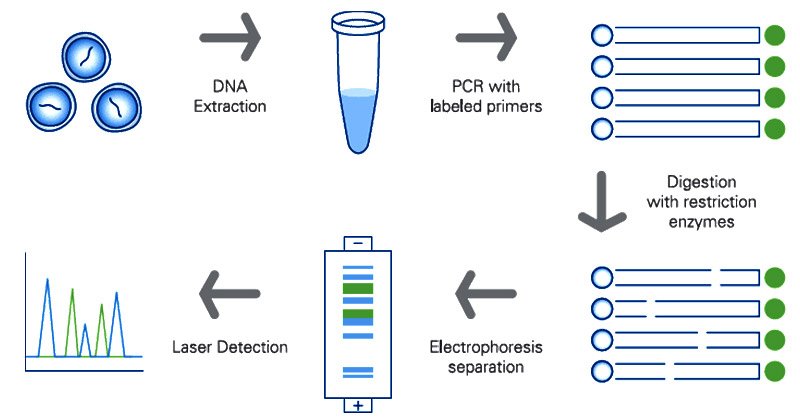
RFLP
- Restriction fragment length polymorphism.
- It is
non-PCR basedtechnique where genomic DNA is digested with restriction enzyme(s). - The whole genetic material is cut at specific nucleotide sequences.
- The digest is put on electrophoreses blotted on a membrane and labelled with probe.
Polymorphismin the hybridization pattern indicates the genetic difference between individuals.
RAPD
- Random amplified polymorphic DNA.
- It is PCR based genome characterization techniques.
- Single short oligonucleotide (9-12 bases) is used as primers for PCR amplification of genomic DNAs.
- Genomic DNA from two different individuals produces different amplification patterns and can be used for molecular characterization of individuals.
DAF
- DNA amplification fingerprinting.
- It is similar to RAPD with difference that primer is shorter of 5-8 nucleotide sequence and used in higher concentration.
AP-PCR
- Arbitrarily primed polymerase chain reaction.
- These patterns are generated by employing single primers of 10-50 nucleotide bases length in PCR of genomic DNA.
AFLP
- Amplified fragment length polymorphism.
- This method is based on PCR amplification of genomic restriction fragments generated by specific restriction enzymes and oligonucleotide adapters of few nucleotide bases.
- DNA is cut with restriction enzymes and double standard oligonudeotide adapters are joined to the ends of DNA fragments.
- Sets of restriction are selectively amplified by 32P labeled primers.
- The amplified products are separated on gel and screened using autoradiography.
SSR
- Simple sequence repeats.
- This is also known as microsatellites.
- These are tandemly arranged repeats of mono, di, tri, tetra penta nucleotides with different lengths of repeat motifs.
STMS
- Sequence Tagged Micro-Satellite Sites.
SNP
- Single or Simple Nucleotide Polymorphism
Selection of Ideal Molecular Markers
- Highly polymorphic nature: It must be polymorphic as it is polymorphism that is measured for genetic diversity studies.
- Co-dominant inheritance: Determination of homozygous and heterozygous states of diploid organisms.
- Frequent occurrence in genome: A marker should be evenly and frequently distributed throughout the genome.
- Selective neutral behaviors: The DNA sequences of any organism are neutral to environmental conditions or management practices.
- Easy Access (Availability): It should be easy, fast and cheap to detect.
- Easy and fast assay
- High reproducibility
Uses of DNA markers
- DNA fingerprinting:
- To establish distinctness among biological entities
- Genetic diversity studies
- Evolutionary studies
- Molecular mapping:
- To prepare saturated genetic map
- Chromosome identification
- Map based cloning of genes
- Marker Assisted Selection
- QTLs
- Disease resistance
- Construction of genetic maps
Use of Molecular Markers in Breeding
Marker Assisted Selection (MAS)
- Marker aided selection is a tool for breeding, wherein genetic marker(s) tightly linked with the desired trait/gene(s) are utilized for indirect selection for that trait in segregating/non-segregating generations. In its simplest form it can be applied to replace evaluation of a trait that is difficult or expensive to evaluate.
- MAS is most useful for traits that are difficult to select e.g., disease resistance, salt tolerance, drought tolerance, heat tolerance, quality traits (aroma of basmati rice, flavour of vegetables).
QTL mapping
- Some of the most difficult tasks of plant breeders relate to the improvement of traits that show a continuous range of values.
- Genetic factors that are responsible for a part of the observed phenotypic variation for a quantitative trait are called quantitative trait loci (QTLs).
- The term QTL was coined by Gelderman.
Tagging of disease resistance genes
- The identification of molecular markers closely linked with resistance genes can facilitate expeditious pyramiding of major genes into elite background, making it more cost effective.
Tagging of male sterility genes
- A cytoplasmic male sterile system is desirable for use in hybrid seed production, as it eliminates the need for hand emasculation.
Diversity evaluation
- Recent advances in molecular biology have provided powerful genetic tools, which can provide rapid and detailed genetic resolution.
- Molecular marker based genotyping involves the development of marker profile unique to an individual.
- This unambiguous pattern of crop varieties obtained using DNA a marker is termed as “DNA Fingerprinting”.
- The technique was developed by
Alec Jefferyin 1985 in human and was used first time in crop (rice) in 1988 by Dallas for cultivar identification.
Heterosis breeding
- Another important application of DNA markers is their prediction of heterosis in hybrids.
- Evaluation of hybrids for heterosis or combining ability in field is expensive.
- Molecular markers have been used to correlate genetic diversity and heterosis in several cereal crops (like rice, oat, and wheat).
Hybrid seed purity testing
- To determine the hybrid seed quality it is to be verified that the designated cross has occurred, the number of self-pollination between the female parents meet the necessary purity and the product has adequate quality.
- For years, the only method to check the hybrid seed purity has been the grow out test. Now the RAPD and RFLP are markers are used to test the purity of F1 hybrids.
Pyramiding of Bt genes
- Many Bt strains with novel insecticidal genes have been found. A desired combination of Cry proteins can be assembled via site-specific recombination vectors into a recipient Bt strain to create a genetically improved biopesticide.
Map based cloning of genes
Gene pyramiding
- Gene pyramiding or stacking can be defined as a process of combining two or more genes from multiple parents to develop elite lines and varieties.
- Pyramiding entails stacking multiple genes leading to the simultaneous expression of more than one gene in a variety.
Genomics
- It is science of identifying the sequence of DNA in species. They are complete set of chromosomes found in the gamete of true diploid (True diploid are individual with two set of chromosomes).
- The genome of
rice,soybean&sorghumare completely sequenced but for wheat it not completely sequenced. - Germplasm is genetic background of a species.
- Gene Cluster is a group of adjacent genes that are identical or related.
- Map unit: The distance between two genes that recombine with a frequency of 1 per cent.
- Cybrids or Cytoplasmic hybrids: Hybrids produced using nucleus of one parent cell and cytoplasm of both the cell are called Cybrids.
- It involves in the fusion of a normal protoplast of one species with protoplast having inactivated nucleus of another species.
- DNA replicase: DNA-synthesizing enzyme required specifically for replication.
- Genetic map is map of genome showing relative positions of genes and/or markers on chromosomes.
- Vector: A plasmid, phage, virus or bacterium used to deliver selected foreign DNA for cloning and in gene transfer.
- Cosmids are plasmid vectors that contain a bacteriophage DNA into phage particles.
- Transgenics plants are genetically modified organisms.
- Transgenics or Genetic Modified Organisms (GMO) are organisms with gene or genetic construct introduced by molecular or recombinant DNA.
- Transformation is a process by which genetic material carried by a species cell is altered by incorporation of exogenous DNA in genome.
- Transcription is process during which the information in a sequence of DNA is used to construct m-RNA.
- Virus that live in bacteria — Bacteriophage (1st observed by Ruska in 1940).
- Transgenic are genetically modified plants, which have been created mobilizing genes of interest from one species to another species.
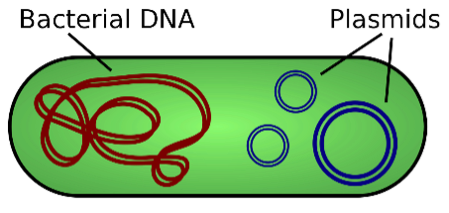
Plasmid
- Plasmid is an independent, double standard, closed circular, self-replicating autonomous molecule of DNA, existing in cells as extra-chromosomal genomes or units, i.e. PBR322, pACYC, Psc101, ColE1 etc.
- Categories of Plasmids
- Stringent plasmids, which replicate only when the chromosome replicates.
- Relaxed plasmids, replicate on their own.
- Ti plasmid: Tumor-induced plasmid, often responsible for crown gall (tumor) induction in plants. Ti plasmids are used as vectors to introduce foreign DNA into plant cells.
- A genetic marker is a gene or DNA sequence with a known location on a chromosome that can be used to identify individuals or species.
- It can be described as a variation (which may arise due to mutation or alteration in the genomic loci) that can be observed.
- Generally, they do not represent the target genes themselves but act as ‘signs’ or ‘flags’.
- Genetic markers that are located in close proximity to genes (i.e. tightly linked) may be referred to as gene ‘tags’.
- Such markers themselves do not affect the phenotype of the trait of interest because they are located only near or ’linked’ to genes controlling the trait.
- All genetic markers occupy specific genomic positions within chromosomes (like genes) called ‘loci’ (singular …