⼍ Cell Coverings
Learn about Cell Memeberane, Cell Wall, Protoplasm
Plasma membrane/Cell membrane/Plasma lemma
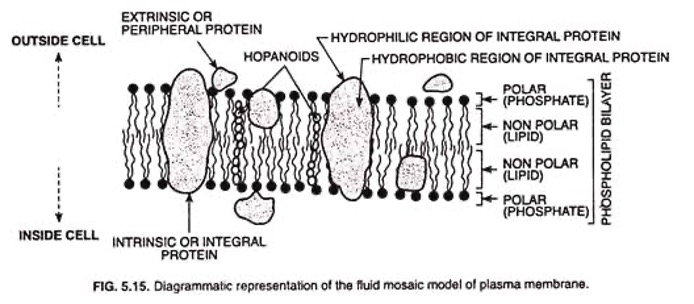
- It is the extremely delicate, thin, elastic and living membrane which surrounds a cell.
- In plant cell, it is present on the inner side of the cell wall. It is made up of two layers of lipid (fat) molecules (size 35 A°) with protein molecules (each 20 A°) sandwiching it and embedded in it.
- It is a
lipo-proteinmembrane.- Protein (20 A° thickness)
- Lipid, bilayer (35 A°)
- Protein (20 A°) monolayer
- Total thickness of the membrane is 75 A° - 100 A°
- Plasma membrane is a selectively permeable (semi-permeable) membrane and its function is osmoregulation of molecules. It protects the internal structures and gives shape and rigidity to cell.
- In animal it is the outermost structure of the cell and hence called `ectoplast’.
- Plasma membrane is absent in virus.
- Sialic acid is a constituent of the cell membrane. It acts as a cell receptor. It is a monosaccharide with a nine-carbon atom.
- Membrane less Cell Organelles: Ribosome, Centriole, Centrosome, Microtubules.
- Single Membrane bound Cell Organelles:
Peroxisomes,Lysosomes,Sphaerosome,Glyoxysomes - Double Membrane bound Cell Organelles:
Nucleus,Mitochondria,Chloroplast
Cell Wall
- Plant cells have an additional protective wall outside the plasma membrane, called the cell wall.
- Cell wall is the
non-livingand thick envelope and is the most important character of the plant cells. - Cell wall is
absent in animal cells. - It is permeable and made up of
cellulose(a type of carbohydrate). - Middle lamella joins primary cell wall of the adjacent cell walls.
- Chemically (middle Lamella)
- Rich in
pectin(cementing material and also used in jelly making). - Ca2+ (trace of Mg also): provides rigidity.
- Rich in
- Primary cell wall contains:
- Cellulose (Polymer of hexose (6c) glucose)
- Hemicellulose (Polymer of mannose/galactose/ Pentose)
- Pectin (hydrophilic)
- Pectin is in primary cell wall in lower plants but in higher plants it is present in middle lamella.
- Plant cell type on the basis of the nature of cell wall: secondary cell wall absent or very thin e.g.
Parenchyma. - Primary wall (cellulose) present + Secondary wall (cellulose only) also present e.g.
collenchyma. - Mechanical property of collenchyma is due to cellulose in its secondary wall.
- Primary wall present + Sec. wall thick (cellulose + lignin) e.g.
Sclerenchyma,Vessels,Tracheids.
- Plant cell type on the basis of the nature of cell wall: secondary cell wall absent or very thin e.g.
- Lignin: Chemically it is
coniferyl alcoholand solid at room temperature. It gives mechanical strength. - Hardness of woody tissue is due to lignin. IBPS AFO 2012
- Plant cell type on the basis of lignin:
- Non-lignified cells (lignin absent): e.g. parenchyma, collenchyma, Sieve tubes.
- Lignified cells (Lignin present): e.g. Sclerenchyma, vessels & tracheids. Classroom stain for lignified cells:
Safranin.
Protoplasm
- All the components of a cell including the cell membrane is called protoplasm.
- It is the
living partof the cell. - The term ‘protoplasm’ was given by
J.E. Purkinje(1830-37). - ‘Plasm’ means substance.
- According to Huxley: Protoplasm is the
physical basis of life. - 80-90 % of protoplasm is water.
- Protoplasm is colloidal in nature and heterogeneous.
- In dry protoplasm, protein is the most abundant constituent (60-70%).
- Protoplasm is a polyphasic colloidal system.
Nucleus
- Nucleus word derived from
Latinword “Kernel”. - Discovery of nucleus:
Robert Brown(1833). - The nucleus is surrounded by a double membrane nuclear envelope (having pores) called nuclear membrane which is chemically lipo-protein.
- The size of nuclei: 5 – 25 µ.
- A true nucleus is absent in bacteria and blue-green algae (cyanobacteria) but nuclear materials are present in them.
- Nucleus is absent in
- Matured mammalian RBCs
- Sieve tube cells in phloem
- Xylem cells (Only a hollow tube that transport water & has no organelle)
- Inside the nuclear membrane there is a colorless dense sap called nuclear sap or nucleoplasm or karyolymph or Karyofluid.
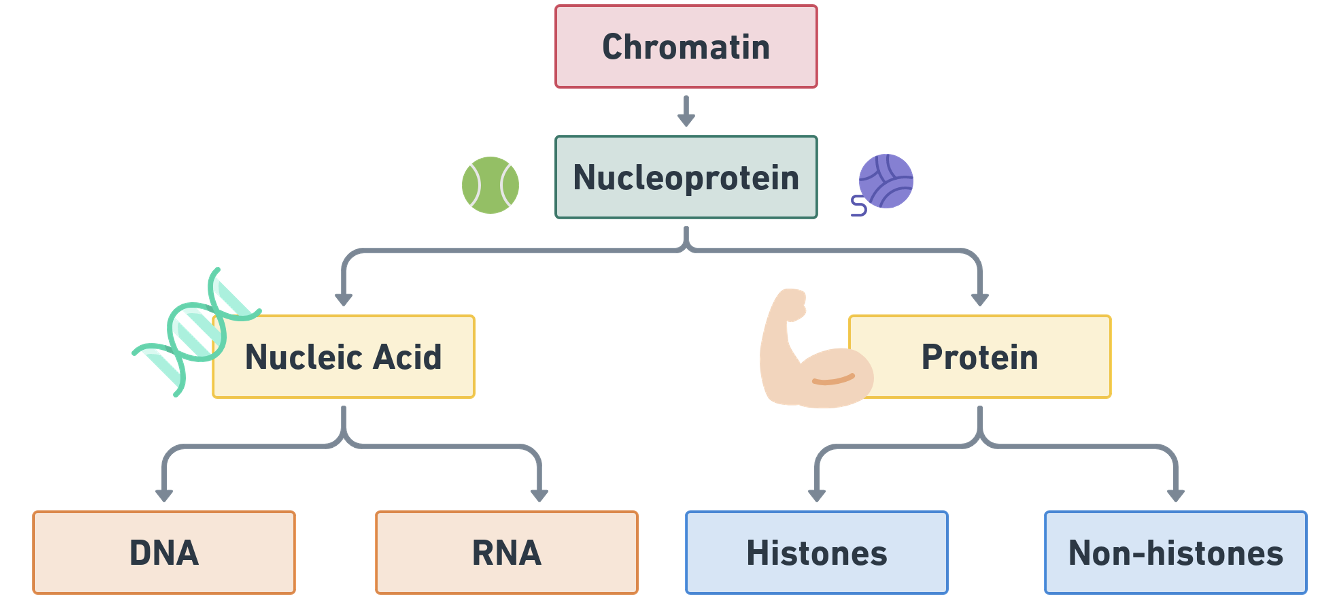
- Inside the nucleoplasm, a tangled mass of threadlike structures is called chromatin.
- Chromatin: made up of DNA + Protein.
- DNA: Deoxyribonucleic acid
- When chromatin condense into rod like bodies during cell division called chromosomes.
- Chromosome contains stretches of DNA. DNA carry information for protein synthesis. The stretches of DNA are called
genes. Genes are passed from parents to the progeny. Hence gene is called hereditary unit and DNA is called hereditary material. Genes control the production of enzymes which guide metabolic activities.
Nucleolus
- A spheroidal and densest organelle in the nucleus is nucleolus.
- Nucleolus contains DNA which synthesizes
ribosomal RNA(Ribonucleic acid). - It is discovered by Fontana (1781). It is rich in RNA but also contains DNA.
- The main function of nucleolus is to synthesize Ribosomal RNA (r RNA). It is attached to the specific site of chromosome which is called ‘nucleolar organizer’.
- In eukaryotes, at least 3 RNA polymerases are present in nucleus viz.
- RNA Polymerase I or A: located in nucleolus, responsible for
r RNA synthesis - RNA Polymerase II or B found in nucleoplasm, responsible for
Hn RNA synthesis(Heterogeneous nuclear RNA) - RNA Polymerase III or C: found in nucleoplasm, responsible for
synthesis of tRNAs or sRNA
- RNA Polymerase I or A: located in nucleolus, responsible for
Chromosome
- Chromosome was first seen by
Strasburgerin 1875 as fine thread. (Discovered) - But the name ‘Chromosome’ (Chroma = colour + soma = body) was first used by
Waldeyerin 1888 using basic dye. - Chromosomes are the carriers of hereditary units i.e. genes. (Given by Morgan) UPPSC 2021
- The chromosome having identical allels for a particular trait is known as
Homozygous. RRB SO 2021
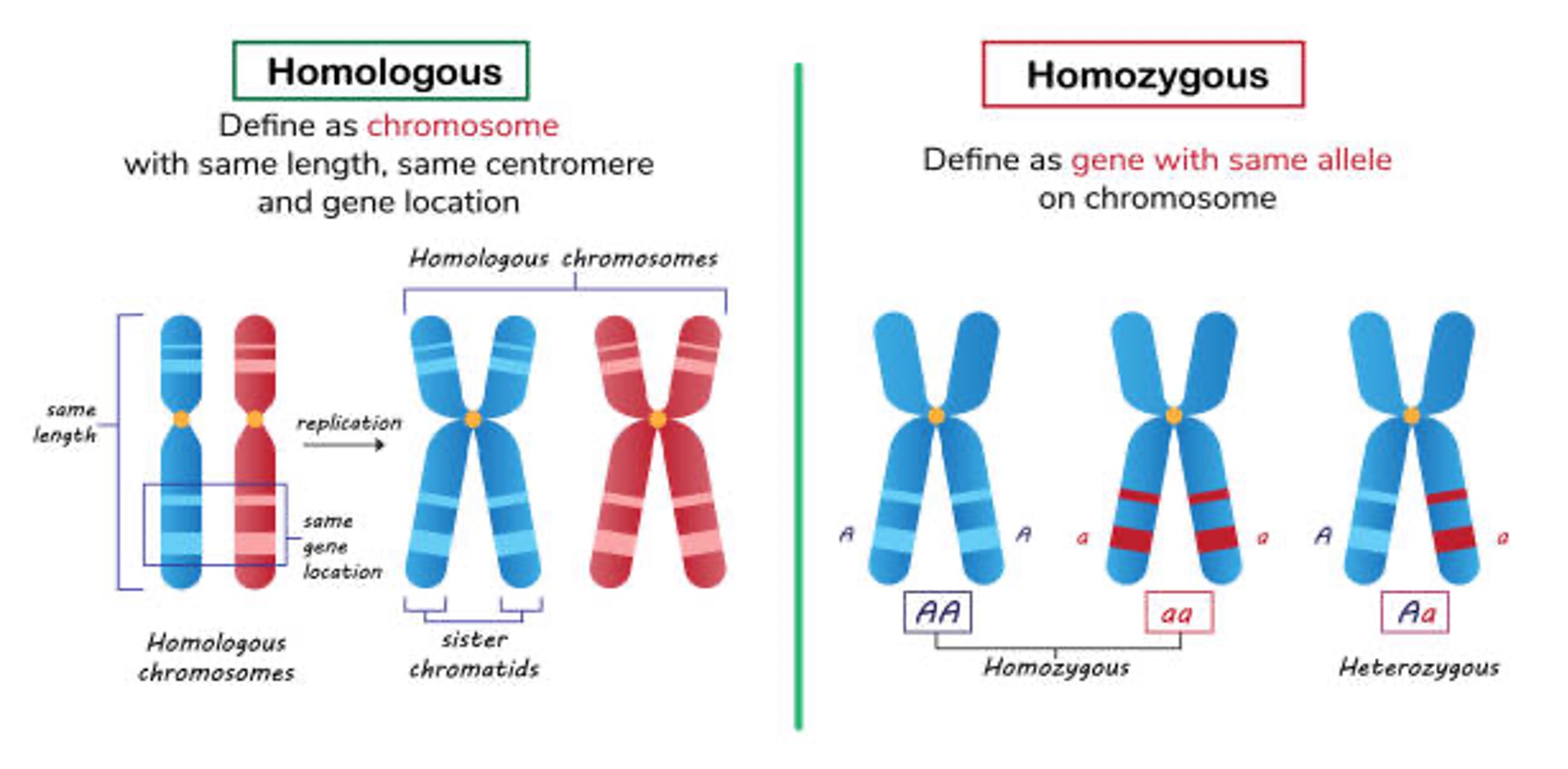
- In all the species, there is a certain no. of chromosomes viz. in human 2n = 46, in rice 2n = 24.
- In Prokaryotes, there is a single chromosome which is circular in shape and is called
genophore(without histone) whereas in eukaryotes chromosomes are rod shaped.
- Nucleic acid is the polymer of nucleotides.
- One nucleotide = Sugar + Nitrogenous base + Phosphate (H3PO4)
- One Nucleoside = Sugar + base only
- Nucleotide = Nucleoside + Phosphoric acid
- Nitrogenous bases are:
- A → Adenine
- G → Guanine
- C → Cytosine
- T → Thymine
- U → Uracil
- Such bases are ringed structure.
- Purines: Two rings e.g. A, G
- Pyrimidines: Single ring e.g. T, U, C
- J.D. Watson & F.H.C. Crick (1953): proposed a model of DNA comprising of two helically intertwined chains tied together by hydrogen bonds, between the purines & pyrimidines called Double Helix Model of DNA.
- Wilkins: X-ray diffraction of DNA.
- In 1962 Watson, Crick & Wilkins got a Nobel Prize for their studies on the structure of DNA.
Mitochondria
- ‘Power house of cell’.
- Energy is generated as
ATP. ATP is known as ‘Energy currency’. - Mitochondria was firstly identified by
Altmanin 1886 as `Bioplast’ and suggested their association with respiration. - The term ‘mitochondria’ has been given by
C. Benda(1898).
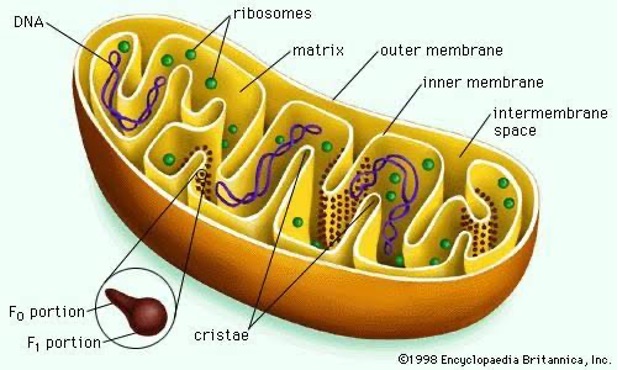
- Matrix of Mitochondria is the site of
aerobic respirationorKrebs cycle. - Site of glycolysis:
Hyaloplasm(not cytoplasm) - Electron transport: Oxysome/F1 particle/Elementary particle.
- Enzymes confined in Peri-mitochondrial space and reactions on inner membrane.
- Mitochondria contain DNA, RNA (0.02% DNA, 3-4% RNA) & ribosomes.
- Hence called ‘semi-autonomous’ body of the cell because of capability of some protein synthesis.
Plastids
- According to Schimper (1885)
- Green coloured plastids are called
chloroplasts - Red, Yellow, Brown, Orange →
Chromoplast(Chromo means Coloured) - Colourless:
Leucoplasts. The main Function of leucoplasts is food storage.
- Green coloured plastids are called
Leucoplast
- Amyloplast: Storage of
Starch - Elaioplast: Storage of
Oils - Aleuronoplast or Proteinoplast: Storage of
Protein
Chlorophyll
- Chloroplast contains pigment chlorophyll
- Chlorophyll a: Blue black, C55H72O5N4Mg
- Chlorophyll b: Green black, C55H70O6N4Mg
- Carotene: Yellowish Orange, C40 H56
- Xanthophyll: Yellow, C40 H56 O2
- Chl. a & Chl. b in green plants: 65%
- Xanthophyll → Yellow, 29%
- Carotene → Red, 6%
- Carotene & Xanthophyll are together called
carotenoid pigment. - In chromoplast, only carotenoid pigments are found.
Plant pigments
- Plastid pigments e.g. chlorophyll, carotenoids; such pigments are soluble in organic solutions only. Found in leaves & skin of fruits.
- Sap pigments: Solution of salts & sugars found in vacuoles e.g.
Anthocyanin(soluble in water) found in flower petals & beets.
Chloroplasts
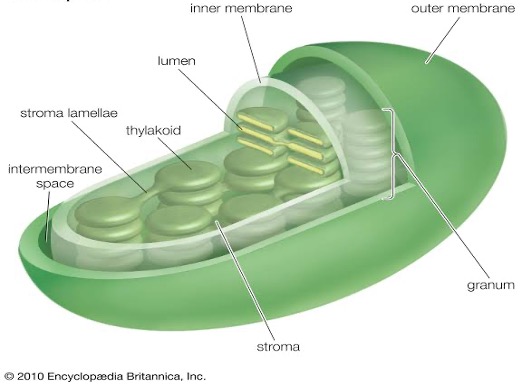
Grana lamellae
- Thylakoids → Quantasomes contain pigment
- Lamellae means membranous tubes.
- Site of Light reaction: Grana/thylacoids/quantasomes → Chlorophyll
- Site of Dark reaction: Stroma/matrix →
RuBISCO - Chloroplast contains DNA (0.5%), RNA (3-4%) and ribosomes (70s). Therefore chloroplast is capable of some independent protein synthesis hence called semi-autonomous body of the cell.
- A plastid has two distinct regions viz. grana & stroma.
- Grans are stacks of membrane bound, flattened, discoid sacs containing chlorophyll molecules. Such molecules are responsible for the food production, hence called the kitchen of the cell.
- The homogenous matrix in which grana are embedded is known as stroma.
Endoplasmic Reticulum (ER)
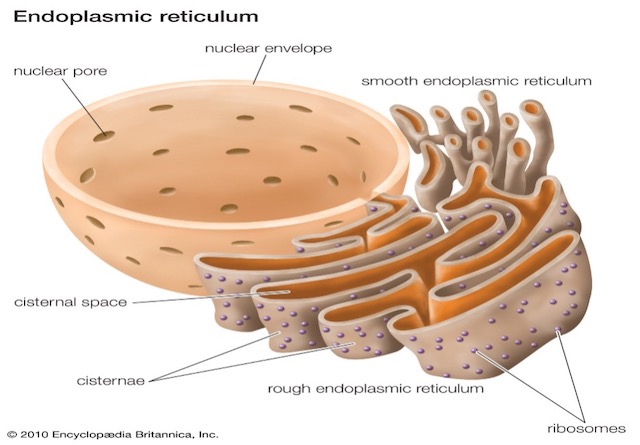
- Endoplasmic Reticulum is a dense network of double membrane (unit membrane) structures running through the cytoplasm.
- It may be continuous, some parts are connected to the nuclear membrane while others are connected to the plasma membrane especially in animals.
- Its origin is from nuclear membranes.
- Ultrastructure of ER was first reported by Porter (1948).
- It is not a stable structure.
- It is capable of being broken down and reconstructed.
- It undergoes partial fragmentation at the time of cell division.
- Two types of ER
- Rough ER (RER): When ribosomes are attached on it.
- Smooth ER (SER): No ribosomes are attached on its surface.
Function
- ER forms endoskeleton to provide a particular shape to the cell (mechanical support), especially in animals.
- Membranes of ER provide the surface for the increased metabolic reaction.
- Intracellular transport of metabolic products or molecules (e.g. protein).
- It helps in the formation of cell plate and nuclear membrane during cell division.
- Synthesis of protein on ribosomes means Rough ER is associated with the protein synthesis.
- Smooth ER (SER) secretes lipids which along with proteins constitute cell membrane by a process called membrane biogenesis.
- Transmission of impulses in animals.
- SER plays a crucial role in detoxifying many poisons & drugs.
Ribosomes/RNA particles
- Consists of r-RNA (ribosomal RNA) 60-40% & Protein 40-60%.
- First observed by Claude (1943) but reported as ‘microsomes’ (ER fragments + ribosomes). Palade (1956) isolated the ribosomes and reported their detailed ultra-structure.
- The term ‘ribosomes’ by R.B. Robert (1958).
Types of RNA (non-genetic)**
- m-RNA (messenger): 5-10% of total RNA
- t-RNA (transfer) / soluble RNA (sRNA): 10-15% of total RNA, Smallest in size.
- Clover leaf model.
- m-RNA and t-RNA directly take part in protein synthesis.
- r-RNA: provides site for protein synthesis.
- 80% of total RNA,
Most stable RNA.
- 80% of total RNA,
- Two major steps are involved in protein synthesis:
Transcriptioni.e. transfer of genetic information from DNA to mRNA.Translationi.e. translation of the language of nucleic acids into that of proteins.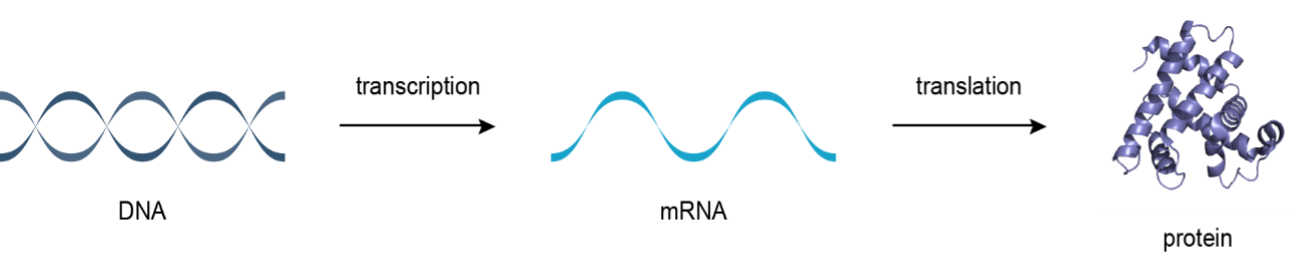
- A ribosome has two subunits, one is smaller and other is bigger.
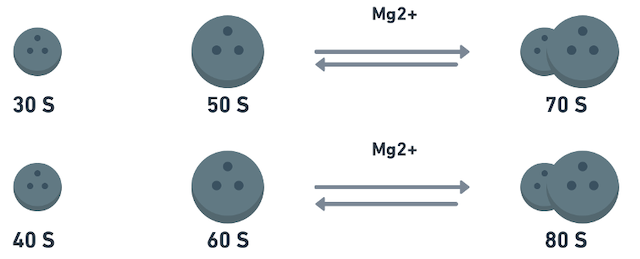
- In bacterial cells & chloroplasts of higher plants, sedimentation coefficient (S) of complete ribosome is
70 Swhich has subunits 50 S and 30 S. - In higher organisms sedimentation coefficient of ribosomes is
80 Swhich has subunits 60 S & 40 S.- Higher concentration of Mg++ ion promotes association of subunits into complete ribosome.
- Lower concentration of Mg++ dissociates these subunits.
- There is no lipid content in ribosomes.
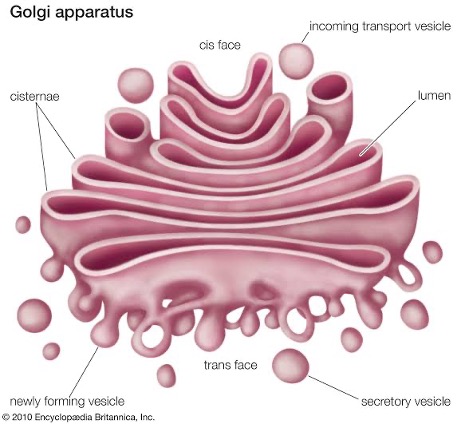
Golgibody/Dictyosome
- Discovered by Camillo Golgi (1898).
- Golgi complex or bodies are formed by stacks of flattened (saucer-shaped) membranes.
- Flattened sacks are called cisternae. Golgi bodies are usually called dictyosomes in plants.
- Its origin from ER.
- ‘Acrosomes’ are associated with Golgi complex.
Functions
- They store, modify, package and condense the protein synthesized in the ribosomes.
- They form the cell plate during cell division.
- They add sugars to some proteins and synthesize some polysaccharides for the cell membrane.
- They set aside digestive enzymes in tiny membrane bound vesicles which become ‘lysosomes’.
Lysosome
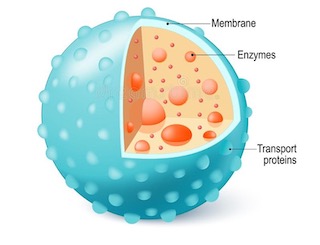
- Lysis means digestion; Soma means body.
- The saclike, small, spherical, single membrane bound vesicles containing digestive enzymes are called lysosomes.
- These enzymes are synthesized in RER which are brought to the ‘Golgi Complex’.
- Lysosomes are formed directly from Golgi complex & indirectly from ER.
- Lysosomes are discovered by De Duve (1955);
mainly found in animalsbut also in some plants like Neurospora. - In animals, epithelial cells of the intestine, kidney cells rich in lysosomes.
Functions
- Intracellular digestion. They help in breaking down (digesting) large molecules of cell.
- They work in defense against bacteria & virus.
- During starvation, lysosomes act on their own cellular organelles & digest them. This result in cell death hence are called ‘
suicidal bag’ or demolition squads means cell autolysis or autophagy.
Spherosomes
- Single membrane bounded mainly in plants.
- Its function is
fat metabolism.
Microsome
- Microsomes are the structures formed when cells are broken up in the laboratory.
- Differential centrifugation can be used to separate them from other cellular debris.
- They are used to imitate the activity of the endoplasmic reticulum in a test tube.
- They are also used to perform experiments that require protein synthesis on a membrane thus aiding in understanding the process of protein formation on the endoplasmic reticulum.
Vacuole
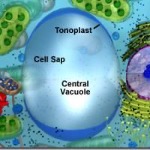
- Important structure in
plant cell. - They may occupy upto 90% of the cell volume.
- Single membrane bound and contain cell sap.
- Function: Osmo-regulation of molecules.
Tonoplastis the membrane of vacuole.
Plasmodesmata
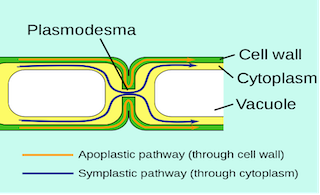
- Found in
plantsonly. - First observed and named by Strasburger (1903) in the form of cytoplasmic strands that connects the protoplast of adjacent cell.
- Origin: from ER
- Function: Provide metabolic contacts between cells e.g. companion cells.
Centrosome
- Present near nucleus,
present in all animal cellsand also in Chlamydomonas, some fungi, Gymnosperms etc. - Two centrioles in one centrosome.
- At the time of mitosis, move to opposite pole and produce astral rays.
- The microtubules of the spindle originate from centrosome during mitosis in both plant and animal cells.
Ergastic Substances
- Non-living cell inclusions e.g. starch, sugar, organic acid, fats, oils, pigments etc.
Genetic Material
- Genetic material DNA & RNA are chemically called nucleic acids. Genetic materials are DNA & RNA
- DNA: Deoxyribose Nucleic Acid
- RNA: Ribose Nucleic Acid
- Genetic material in most of the organism is DNA.
- ‘DNA’ is the genetic material and not the protein’ was described by O.T. Avery, C.H. Macleod and M. Mc Carty on E. coli in 1944.
- Nucleic acids were first isolated by Miescher (1868) from the nuclei of white blood cells (WBC) of pus.
- First time In vitro synthesis of DNA:
A. Kornberg - In vitro synthesis of RNA:
S. Ochoa - Artificial Synthesis of gene which coded for alanine transfer RNA (t RNA) from yeast:
H.G. Khorana&K.L. Agrawal. - One gene - One enzyme hypothesis was given by
Beadle & Tatumin 1943 while working on biochemical, mutant Neorospora crassa (fungus). - How gene operates? i.e. operon concept of gene was given by
Jacob & Monod. - Modern concept of gene i.e. gene fine structure was given by
Benzer - According to the classical concept, the smallest unit for recombination, mutation & function was only gene. But Later on three terms have been coined by Benzer viz.
Reconis the smallest unit for recombination. It consists of maximum of two pairs of nucleotides & may be only one.Muton: It is the smallest unit of mutation. An alternation in a single nucleotide pair can result in the mutation. Muton is thesmallestin size.Cistron: It is the functional unit where hundreds of nucleotide pairs take part. It is thelargestin size. Generally we use ‘gene’ instead of cistron.
- One gene controls one character
- One gene → One protein
- One gene → One chain
- One gene → One-polypeptide
- One cistrone → One polypeptide
Structure of DNA
- According to J.D. Watson & F.H.C. Crick (
1953) - DNA molecule has two polynucleotide chains which are wrapped helically around each other in such a way that sugar-phosphate chain is on the outside and purine (A, G) pyrimidine (C, T) on the inside of the helix.
- E. Chargaff & et. al observed that:
- The concentration of thymine was always equal to the concentration of adenine and the conc. of cytosine was always equal to the conc. of guanine.
- The total conc. of Purines (A + G) was always equal to the total conc. of Pyrimidine (T+C)
- But the ratio of (A+T)/(C+G) called
Base pair ratiowas found to vary widely in DNAs of different species. - The two polynucleotide strands are held together by hydrogen bonds between specific pairs of purines & pyrimidines.
- Adenine is always paired with Thymine by two hydrogen bonds and Guanine is always paired with Cytosine by three H-bonds.
- The base pairs are stacked
3.4 A°apart with10 base pairsper turn (360°) of the double helix.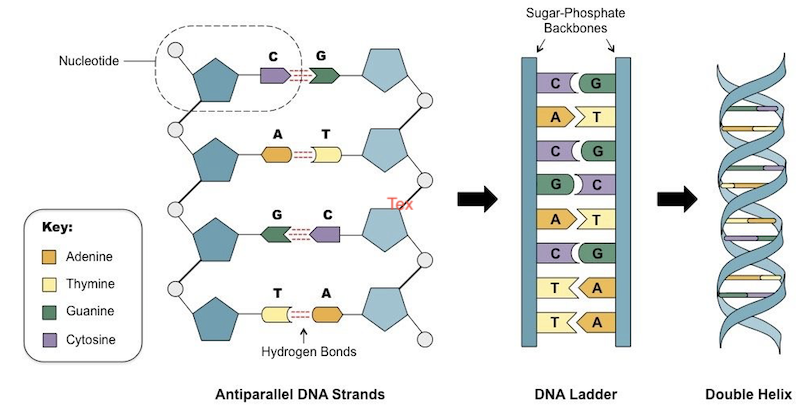
- Once the sequence of bases in one strand is known, the sequence of bases in the other strand is also known because of the specific base-pairing.
- The two strands are thus said to be
complementaryi.e. not identical. Means the sequence of nucleotides in one chain dictates the sequence of nucleotides in the other. The two strands runanti-paralleli.e. have opposite directions. - Both polynucleotides strands are separated by
20 A°distance.
Structure of RNA
- RNA is either single stranded or double stranded but not helical like DNA.
- In most plant viruses, genetic material is RNA. In many bacteriophages, genetic material is RNA.
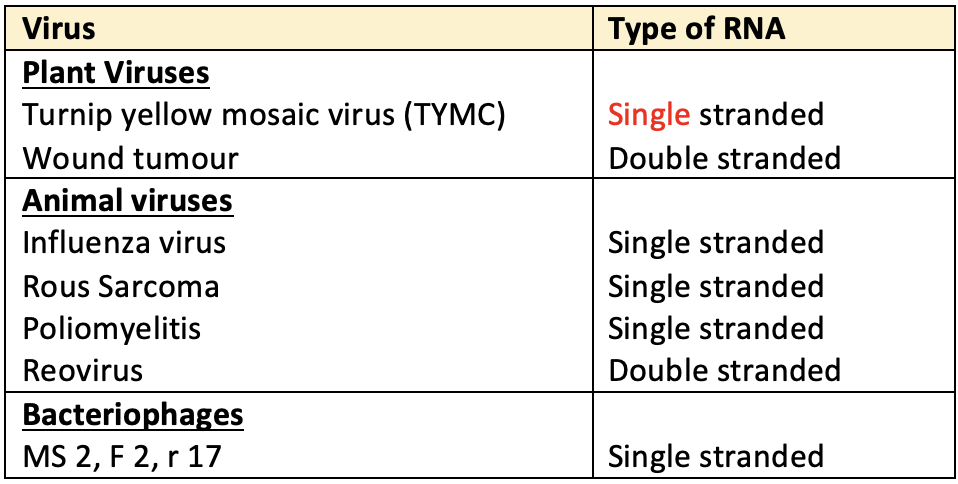
- When RNA is double stranded, it generally follows the same rules of base pairing as in case of DNA.
- The non-genetic RNAs except t RNA (of prokaryotes & eukaryotes) also have single stranded t-RNA (s-RNA is soluble and non-genetic) is double stranded but non-helical.
- Each strand of RNA is polynucleotide. RNA is the genetic material where organisms have only RNA (not DNA) but organisms which have DNA along with RNA, use of the RNA is carrying the orders of DNA and in this way RNA has no genetic role so called non-genetic RNA. Non genetic RNA is synthesized on DNA template. Non genetic RNAs are m-RNA, t-RNA & r-RNA.
- Genetic RNA of viruses is self-replicating i.e. it can-produce its own replica by itself. So its model of replication is called
RNA-dependent RNA synthesis.
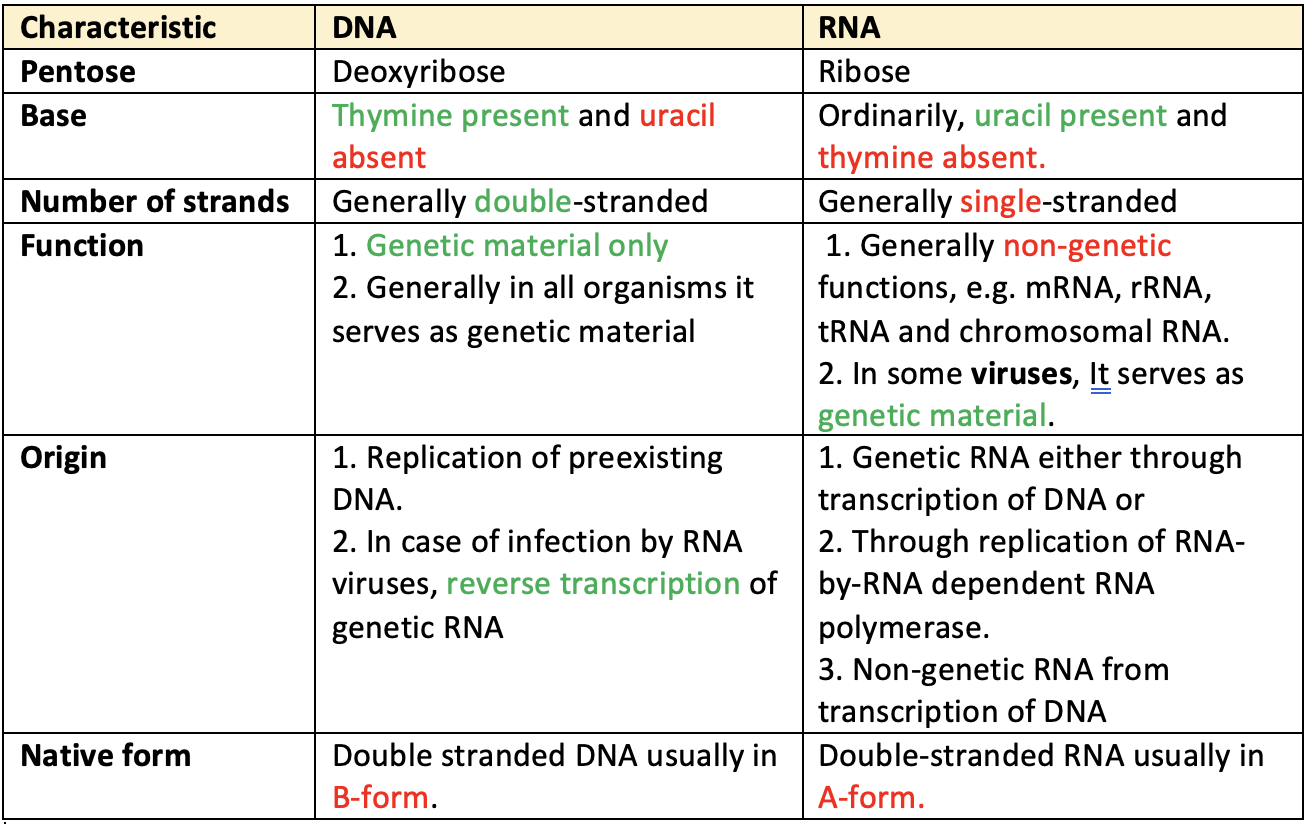
Genetic Code
- R.H. Holley, H.G. Khorana & M.W Nirenberg got Nobel Prize for deciphering the genetic code.
- DNA molecule is the carrier of genetic information. This information is in the form of certain special language of code words which utilizes the four nitrogen bases (e.g. ATCG) of DNA for its symbols. Such coded message is called cryptogram.
- Nucleic acids govern the protein synthesis. Proteins have 20 different amino acids but nucleic acid has only 4 different bases. It means 20 amino acids are the alphabets of the language of Proteins but 4 bases are the alphabets of the language of nucleic acids. Genetic information are passed on to the protein synthesis through mRNA.
- Since 20 amino acids are to be coded, triplet code of bases gives 4x4x4 = 64 codons which are enough.
- Out of 64 triplets, 44 triplets are excess which show that more than one codon are present for the same amino acids.
- An anticodon is a sequence of three nitrogenous bases found on t-RNA.
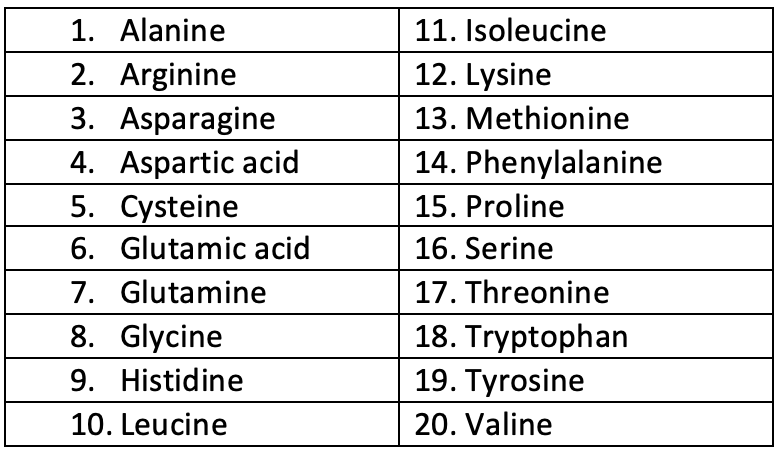
- Primary structure of protein is amino acids. These essential amino acids are
20 in numberviz:
Characteristics of triplet code
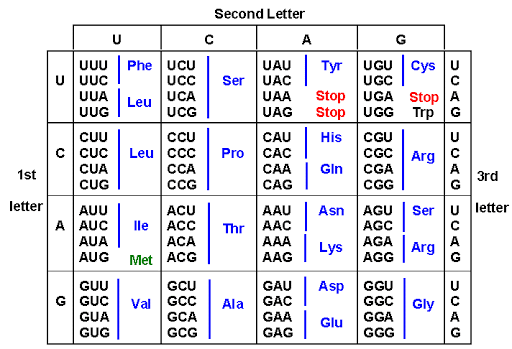
- The code is triplet: Singlet and doublet codes are not enough to code for 20 amino acids. Triplet code is the minimum requirement.
- The code is degenerate: In triplet code for a particular amino acid
more than one word (synonyms)can be used. This is called degenerate code but in a non-degenerate code there would be one to one relationship between amino acids and the codons. So that 44 codons out of 64, will be useless codons e.g. 3 amino acids-arginine, serine & leucine each has six synonymous codons. All codons starting with CC specify proline (CCU, CCC, CCA, CCG). - The code is non-overlapping means a base in an mRNA is not used for two different codons. The same base cannot be used for two different codons for synthesis of the same amino acid. The same base can be used for different codons only at different occasions in time and/ or space.
- The code is comma less. No codon is reserved for punctuation. After the coding of the one amino acid, the second amino acid is automatically coded by the next three letters.
- The code is ambiguous means the same codon may specify more than one amino acid. e.g. UUU codon usually code for phenylalanine but in presence of streptomycin, may also code for isoleucine, leucine or serine.
- The code is universal i.e. the same code applies in all kinds of living systems.
- Starting codons:
AUG codonis called starting or chain initiation codon which initiates the synthesis of polypeptide chain. - Non-sense codons: The
UAA,UGA&UAGcodons do not specify any amino acid. Therefore these codons are called non-sense codons or termination codons. Termination codons terminate the translation of a particular polypeptide.
Plasma membrane/Cell membrane/Plasma lemma

- It is the extremely delicate, thin, elastic and living membrane which surrounds a cell.
- In plant cell, it is present on the inner side of the cell wall. It is made up of two layers of lipid (fat) molecules (size 35 A°) with protein molecules (each 20 A°) sandwiching it and embedded in it.
- It is a
lipo-proteinmembrane.- Protein (20 A° thickness)
- Lipid, bilayer (35 A°)
- Protein (20 A°) monolayer
- Total thickness of the membrane is 75 A° - 100 A°
- Plasma membrane is a selectively permeable (semi-permeable) membrane and its function is osmoregulation of molecules. It protects the internal structures and gives shape and rigidity to cell.
- In animal it is the outermost structure of the cell and hence called `ectoplast’.
- Plasma membrane is absent in …
Become Successful With AgriDots
Learn the essential skills for getting a seat in the Exam with
🦄 You are a pro member!
Only use this page if purchasing a gift or enterprise account
Plan
Rs
- Unlimited access to PRO courses
- Quizzes with hand-picked meme prizes
- Invite to private Discord chat
- Free Sticker emailed
Lifetime
Rs
1,499
once
- All PRO-tier benefits
- Single payment, lifetime access
- 4,200 bonus xp points
- Next Level
T-shirt shipped worldwide

Yo! You just found a 20% discount using 👉 EASTEREGG

High-quality fitted cotton shirt produced by Next Level Apparel